Abstract
Images document scientific discoveries and are prevalent in modern biomedical research. Microscopy imaging in particular is currently undergoing rapid technological advancements. However, for scientists wishing to publish obtained images and image-analysis results, there are currently no unified guidelines for best practices. Consequently, microscopy images and image data in publications may be unclear or difficult to interpret. Here, we present community-developed checklists for preparing light microscopy images and describing image analyses for publications. These checklists offer authors, readers and publishers key recommendations for image formatting and annotation, color selection, data availability and reporting image-analysis workflows. The goal of our guidelines is to increase the clarity and reproducibility of image figures and thereby to heighten the quality and explanatory power of microscopy data.
Similar content being viewed by others
Main
Images and their analyses are widespread in life science and medicine. Microscopy imaging is a dynamic area of technology development, in terms of both hardware and software. This is especially true in light microscopy, with recent improvements enabling sensitive, fast and high-resolution imaging of diverse samples. Existing resources developed by scientists can help researchers to navigate designing microscopy experiments1,2,3 and cover aspects such as sample preparation1, microscope usage1,4, method reporting5,6,7,8 and fluorophore and filter usage9,10. Despite widespread adoption of microscopy as a tool for biology and biomedical research, the resulting image figures in publications at times fail to fully communicate results or are not entirely understandable to audiences. This may be because authors do not include comprehensive imaging method statements11 or because they omit basic information in figures such as specimen size or color legends12, which are key to fully understanding the data. To ensure that images are presented in a clear, standardized, and reproducible manner, it is essential that the scientific community establishes unified and harmonized guidelines for image communication in publications.
Images document biological samples and the ranges of their phenotypes. Increasingly, microscopy images are also a source of quantitative biological data, and variables are measured with a growing number of open source image-analysis software packages such as Fiji–ImageJ13, CellProfiler14, KNIME15 and Python software libraries16 (https://scikit-image.org (ref. 17)) and commercial software packages such as ZEN, LAS X, NIS-Elements, Amira-Avizo, Imaris, Arivis Vision 4D and Huygens18. Image analysis is often a workflow of many steps, such as image reconstruction, segmentation, processing, rendering, visualization and statistical analysis, many of which require expert knowledge19,20. A comprehensive publication of quantitative image data should then include not only basic specimen and imaging information, but also the image-processing and analysis steps that produced the extracted data and statistics. Toward fully reproducible image analysis, it is also essential that images and workflows are available to the community, for example, in image repositories or archives21,22,23, and code repositories such as GitHub24.
To ensure that image figures provide insights to their readership, any supportive experimental metadata and image-analysis workflows must be clear and understandable (‘what is the pixel size?’, ‘what does the arrow mean?’), accessible (‘are colors visible to color-blind audiences?’), representative (no cherry picking) and reproducible (‘how were the data processed?’, ‘can one access and reanalyze the images?’). In the framework of the initiative for ‘Quality Assessment and Reproducibility for Instruments and Images in Light Microscopy’ (QUAREP-LiMi)25,26, the ‘Image Analysis and Visualization Working Group’ established community consensus checklists to help scientists to publish understandable and reproducible light microscopy images and image-analysis procedures. When applicable, the checklists are aligned with the FAIR (findability, accessibility, interoperability and reusability) principles, which were developed as recommendations for research data27.
Scope of checklists
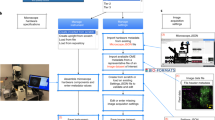
The scope of the checklists is to help scientists to publish fully understandable and interpretable images and results from image analysis (Fig. 1). In this work, the term ‘images’ includes raw or essentially unprocessed light microscope data, compressed or reconstructed images and quantification results obtained through image analysis. While the focus of QUAREP-LiMi is on light microscopy in the life sciences, the principles may also apply to figures with other images (photos, electron micrographs, medical images) and to image data beyond the life sciences. The checklists are intended for all users, from beginners and laypersons who only occasionally work with light microscopy to experienced users and also experts, such as core facility staff or members of the bioimage community, who review image data or teach image processing.
The checklists do not include principles for designing imaging experiments or recommendations to avoid image manipulation. Previous literature covers experimental design for microscopy images, including accurate and reproducible image acquisition and ensuring image quality2,28, examples and recommendations for avoiding misleading images1,29,30,31,32,33, detection of image manipulation34,35,36,37, appropriate image handling and analysis7,19,38,39, guidelines for writing materials and methods sections for images40, and recommendations for general figure preparation41.
The checklists cover image (Fig. 2) and image-analysis (Fig. 3) publication and are structured into three levels that prioritize legibility and reproducibility.
-
The first reporting level (‘minimal’) describes necessary, non-negotiable requirements for the publication of image data (microscopy images, data obtained through image analysis). Scientists can use these minimal criteria to identify crucial gaps before publication.
-
The second reporting level (‘recommended’) defines measures to ensure the understandability of images and aims to reduce efforts toward evaluating image analysis. We encourage scientists to aim for the ‘recommended’ level as their image-publication goal. However, we acknowledge that some aspects (for example, large data in repositories) may be unattainable today for some authors.
-
The third reporting level (‘ideal’) includes recommendations that we encourage scientists to consider adopting in the future.
Checklists for image publication
Image formatting
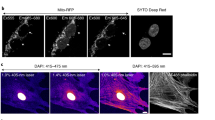
After exploring, processing and analyzing image data, authors then communicate insights in publications with figures as visual evidence. Preparing a figure begins with the selection of representative images from the dataset that illustrate the message. When quantitative measurements are reported in a chart, an example of the input image should be shown; when ranges of phenotypes are described, several images may be necessary to illustrate diversity. To quickly focus the audience on key structures in the image, it is permitted to crop areas without data or with irrelevant data (Fig. 4a). As a rule, cropping, similar to selecting the field of view on the microscope, is allowed, as long as this does not change the meaning of the conveyed image.
a–c, Image formatting may include image cropping, rotation and resizing (a), image spacing in the figure (b) and presenting several magnifications (zoom, inset) of images (c). d–g, Image colors and channels. d, Adjust brightness and/or contrast to achieve good visibility of the imaging signal. e, Channel information should be annotated and visible to audiences (high contrast to background color, visible to color-blind audiences). f, Image details are most unbiased in grayscale. g, It is best practice to publish legends for color intensities (intensity and/or calibration scales) with images, and this is recommended for pseudocolor scales.
Next, specimens are often presented in consensus orientation in figures (for example, apical side of cells upward, tree top upward), which may require image rotation. When such rotation is done in vector software, pixel interpolation is not necessary, as the square representing a pixel can be rotated and size changed as is, preventing any optical data modification. When rotation is done in pixel-based image-processing software, however, any rotation that is not a multiple of 90 degrees changes the intensity values through interpolation and therefore alters the information in the image31,42,43. The effect of interpolation, while it may be negligible in large images composed of many pixels, can greatly distort the information in small or zoomed images composed of fewer than 100 × 100 pixels.
When cropping and rotating, authors should ensure that the operation does not affect the original information contained in the image; while loss in image quality may be acceptable, quantifications, especially intensity measurements, should be performed beforehand39. In a figure, individual images should be well separated (spacing, border; Fig. 4b) to avoid misleading image splicing29,31.
When presenting two magnification views of the same image (for example, a full and a zoomed or inset view), the position of the inset in the full-view image should be made clear; if the inset is placed on top of the full-view image, for example, to save space, it should not obstruct key data (Fig. 4c). If an inset is digitally zoomed, the original pixels should not be interpolated but ‘resized’ to maintain the original resolution. Overall, the image should be sufficient in size so that audiences can identify all relevant details.
Image colors and channels
Fluorescent light microscopes use a range of wavelengths to generate images of specimens. In the images, the light intensity for individual wavelengths, most often in grayscale, is assigned or mapped to a visible color scheme. In multicolored images, several channels are overlaid to compare data from several channels.
Microscopy images often must be processed to adapt the bit depth to the visible range2,44. Usually, brightness and contrast are adjusted for each channel independently in many software platforms (for example, ImageJ–Fiji) by defining the minimum and maximum displayed intensity values before converting these into eight-bit images (for screen display, printing). Intensity-range adjustments should be monitored (for example, with the image histogram) and performed with care: too wide of an intensity range results in ‘faded’ images that lack details, while too narrow of an intensity range removes data (Fig. 4d). Scientists must be especially attentive with auto-contrast and auto-level, image-intensity normalization, non-linear adjustments (‘gamma’, histogram equalization and local contrast, for example, CLAHE45), image filters and image-restoration methods (for example, deconvolution, Noise2Void, CARE46,47,48,49, as their improper application may result in misleading images. When images are quantitatively compared in an experiment, the same adjustments and processing steps must be applied. If deemed critical for understanding the image data, advanced image-processing steps (for example, deconvolution, Noise2Void, CARE) may need to be indicated in the figure or figure legend, in addition to the materials and methods sections.
Next, image colors must be interpretable and accessible to readers and not misleading50. For full-color (for example, histology) images, the staining and preparation methods and, for fluorescence microscope images, channel-specific information (fluorophore or labeled biomolecule) should be annotated (Fig. 4e and Image annotation). In fluorescence microscope images, channels can be assigned a user-defined color scheme, often referred to as a lookup table (LUT), which should be chosen such that the imaged structures are easily distinguishable from the background and accessible to color-blind audiences12. Grayscale color schemes are best for single channels because they are uniformly perceived, allowing unbiased interpretations of intensity values in a given image. Inverting image LUTs to display intensities on a white instead of a black background may enhance signal contrast further, but be aware that different software handles this calculation differently.
A few steps may improve the understandability of colors overall. For multicolored fluorescent images, showing individual channels in separate, grayscale panels provides the best visibility and the highest contrast of detailed structures. Grayscale images are also accessible to all audiences, including color-blind persons (Fig. 4f). If several channels must be merged into one image, choose a color combination visible also to color-blind audiences12,51. A separate, linear-adjusted grayscale version may also help when images are adjusted with non-linear adjustments or pseudocolored LUTs (for example, ‘jet,’ ‘viridis’ and ‘Union Jack’), which map intensity values to a dual- or multiple-color sequential scheme. Annotation of intensity values with an intensity scale bar (‘calibration bar’) helps to orient readers and is essential for pseudocolored and non-linear color schemes (Fig. 4g). Calibration bars should indicate absolute intensity values to inform audiences about the displayed intensity range and can be prepared with Imaris and ImageJ–Fiji (ImageJ user guide: https://imagej.net/ij/docs/guide/).
Image annotation
Image acquisition must be described in detail in the methods section. Additionally, to best communicate image data, some of this information is required or at least beneficial if it is associated with the figure itself as a caption or an annotation. Light microscopy images show objects sized from submicrometer to millimeter resolution. As physical size is not obvious without context, annotating the scale for publication is therefore necessary. A scale bar of a given size (in or next to the image) is needed to orient audiences (Fig. 5a). The corresponding size statement or dimension, for example, ‘0.5 mm’, can be placed next to the scale bar (when not possible in the figure legend). To avoid quality changes (pixelated or illegible text) when adapting (resizing, compressing) figures for publication, annotations should be added as vector graphics. Statements about the physical dimensions of the entire image are acceptable alternatives to scale bars. Magnification statements should be avoided, as pixel size can be determined by a number of factors, for example, sampling rate or binning, and does not depend only on the objective magnification.
a–d, Image annotation. a, Possible ways to provide scale information. b, Features in images can be annotated with symbols, letters or ROI. c,d, For advanced image publication, information on the anatomical view or intervals in time-lapses may be required. hh:mm, time in hours and minutes; 3D, three dimensional. For local storage, size limit is not applicable (N/A). e–g, Image availability. e, Currently, image data are often shared ‘upon request’. f,g, More images along with the image metadata should be available for download in public databases (f) and, in the future, also should be archived in dedicated, added-value databases, in which images are machine searchable or curated (g).
Many images include further annotations such as symbols (arrows, asterisks), letter codes or regions of interest (ROI; dashed shapes), which must be explained in the figure or the figure legend (Fig. 5b). Symbols that resemble image data should be avoided, and note that symbols with clear vertical or horizontal arrangement are easier to distinguish than randomly oriented symbols on busy backgrounds (for example, ref. 12). At times, additional annotations may help readers to interpret the figure when the anatomical section of a three-dimensional object or the imaging frequency for time-lapse data is provided too (Fig. 5c,d). All annotations, including scale bars and ROI indications, must be legible, that is, have sufficient size or point size, line widths and colors in high contrast to the background. In addition to being legible, scale bars should have a meaningful length with regard to the object shown. Annotations placed on top of images should not obscure key image data and should be legible to color-blind persons.
Image availability
Image-processing operations should not overwrite the original microscope image42,43, and, upon publication, both the original image (or a lossless compressed version) and the image file shown in the published figure should be available. The specific file type of the original image depends on the microscope type and the vendor. The definition of ‘original data’ or ‘raw data’ and whether their storage is feasible depend on the specific microscopy technique. In data-heavy techniques (for example, light sheet microscopy) it may be acceptable if the cropped, binned or lossy compressed images, which faithfully capture the key scientific content, are made available. To retain the metadata, a conversion into standard formats or open formats such as OME-TIFF52, which support uncompressed and lossless but also lossy (compressed) files, is compatible with broad applications to allow reanalysis of image data. If only a compressed version may be kept (that is, a file in which image channels and annotations are irretrievably merged), PNG files are superior to the JPEG format, as they allow lossless compression43.
As a minimal requirement, the image files shown in figures or used for quantification should be available. When possible (see limitations above), lossless compressed files, which allow replication of the analysis workflow, should be shared or made available (Fig. 5e–g). We strongly discourage author statements that images ‘are available upon request’, as this has been shown to be inefficient53,54; however, at present, infrastructure is not sufficiently in place to ban this option. A clear advancement is depositing both the published and the original images in a public repository with an open license that permits reuse in the scientific context (CC-BY, CC0). Zenodo, Open Science Framework and figshare are currently essentially free options also for image data; however, some of these have file size limitations (Fig. 5e–g and Extended Data Fig. 1). OMERO servers (https://www.openmicroscopy.org/omero/institution/) enable institutions but also individual laboratories to host public or private (access-controlled) image-sharing databases (see Extended Data Fig. 1 for an overview of current repositories). In the long term (‘ideal’), uploading images with all experimental metadata to dedicated, specialized or fully searchable image databases has the potential to unlock the full power of image data for automated image and/or metadata searches and the possibility of image data reuse. Databases that allow such functionalities and more include the BioImage Archive (a primary repository that accepts any image data in publications; https://www.ebi.ac.uk/bioimage-archive/), the Image Data Resource (which publishes specific reference image datasets; https://idr.openmicroscopy.org/) and EMPIAR (a dedicated resource for electron microscopy datasets; https://www.ebi.ac.uk/empiar/). At present, most of the available options are free of charge for 20–50-GB datasets; however, dedicated image databases have strict requirements regarding file type and metadata (Fig. 5e–g and Extended Data Fig. 1). To be inclusive, we do not enforce the use of online repositories but require as a minimal measure that scientists are prepared to share image data. Costly and expert-driven storage solutions are at present not accessible to all scientists in the global imaging community.
Checklists for publication of image-analysis workflows
Image-analysis workflows usually combine several processing steps carried out in a specific sequence to mathematically transform the input image data into a result (that is, image for visualization or data for a plot; Fig. 6 and ref. 39). As images are numerical data, image processing invariably changes these data and thus needs to be transparently documented31,39,43. We developed separate checklists for scientists wishing to publish results originating from image-processing and image-analysis workflows (Fig. 3). Focusing on easy implementation of the checklists, we propose three categories:
-
1.
Established workflows or workflow templates: workflows available in the scientific literature or well established in the respective fields.
-
2.
New workflows: established or new image-analysis components (available in software platforms or libraries) are assembled by researchers into a new workflow.
-
3.
Machine learning workflows: machine learning uses an extended technical terminology, and machine learning workflows that use deep neural networks (‘deep learning’) face unique challenges with respect to reproducibility. Given the rapid advancements in this field, we created a separate machine learning checklist.
a, An established workflow template is applied on new image data to produce a result (plot). b, A new sequence of existing image-analysis components is assembled into a new workflow for a specific analysis (image segmentation). c, Machine learning workflows learn specific tasks from data, and the resulting model is applied to obtain results.
Established workflows
Examples of well-established workflows are published pipelines for CellProfiler (CellProfiler published pipelines, https://cellprofiler.org/published-pipelines; CellProfiler examples, https://cellprofiler.org/examples), workflows in KNIME55, specialized plugins and easy-to-use scripts in ImageJ56,57,58 and tools and plugins that solve generic image-analysis problems such as tracking59 or pixel classification60,61. For these workflows, extensive expertise, documentation and tutorials already exist that allow others (for example, reviewers, readers) to reproduce the workflow and to judge the validity of the results. Scientists publishing images or image-analysis results processed with established workflows thus can focus on documenting key parameters only.
Minimal
The authors must cite the used workflow. The specific software platform or library needs to be cited if the workflow is not available as a standalone tool. Key processing parameters must be reported. To validate the performance of the workflow and its settings, example input and output data must be provided. Any manual interventions (for example, ROI) must be documented. To ensure proper reproduction of the workflow, the precise version numbers of the workflow and the software platform used are vital and must be documented in the methods. If the used software does not allow the researcher to easily define and retrieve a specific version number, the exact version used must be deposited as a usable executable or code.
Recommended
Authors should state all settings in the methods or the supplementary material of the article. Providing data upon request is an ineffective method for data sharing54. Thus, authors should provide example input and output data and any manual ROI via a public repository (see Image availability).
Ideal
Documenting the use of software in the form of a screen recording or, in the case of command line tools, by reporting all executed commands in detail greatly facilitates understanding the workflow application and therefore reproduction. To avoid any variation arising from factors such as computer hardware or operating system, authors could provide cloud-hosted solutions62,63,64 (kiosk-imageJ-plugin, https://github.com/vanvalenlab/kiosk-imagej-plugin) or the workflow packaged in a software container (docker, https://www.docker.com/; Singularity, https://docs.sylabs.io/guides/3.5/user-guide/introduction.html)65.
New workflows
New image-analysis workflows assemble components into a new sequence, for example, a macro in Fiji, a pipeline in CellProfiler or a workflow in KNIME in an original way. To ensure reproducibility of the analysis, it is essential to report the specific composition and sequence of such new workflows.
Minimal
The individual components used in the new workflow must be cited, named and/or described in detail in the methods section along with the software platform used. It is essential that scientists specify or provide the exact software versions of the used components and software platform in the methods whenever possible. Authors must describe the sequence in which these components have been applied. Key settings (for example, settings that deviate from default settings) must be documented in the methods section. Finally, the developed workflow must be shared as code (for example, via code repositories such as https://github.com/) or pipelines (for example, KNIME workflow, CellProfiler pipeline). Example input and output and any manually generated inputs (that is, ROI) must be made available (Image availability). For new workflows that were created using software that does not allow scripting, the workflow steps must be carefully described with text.
Recommended
All settings of the workflow must be disclosed and described to help reproduction of the analysis. Example input, output and manual inputs (ROI) should be provided via public repositories such as Zenodo. The developer should describe the rationale as well as the limitations of the workflow and the used components in more detail in the methods or supplementary material. Whenever possible, evidence of the adequacy and efficiency of the used algorithms on the published data and potentially even comparisons to related established workflows facilitate such documentation.
Ideal
To further promote reproducibility, documentation such as a screen recording or a text-based tutorial of the application of the workflow should be added. To enable efficient reproduction of an analysis with a new workflow, authors should provide easy installs (for example, update sites, packages) or easy software reproduction (for example, via software containers) and easy-to-use user interfaces of software (that is, graphical user interfaces). The new workflow should also be published as independent methods papers with extensive documentation and online resources55,56,57,58,59,60,61. In sum, extensive documentation and ease of installation and use will ultimately contribute to the new workflow becoming well established and reproduced within the community (a future established and published workflow template)66.
Machine learning workflows
Machine learning and especially deep learning have recently become capable of surpassing the quality of results of even the most sophisticated conventional algorithms and workflows and are continuing to advance67. Deep learning procedures are quickly adapted to microscopy image tasks such as U-Net68 for cell segmentation69, Noise2Void for image reconstruction47, StarDist70,71 and Cellpose62 for instance segmentation, DeepProfiler72 for feature extraction and Piximi (https://www.piximi.app/) for image classification.
In machine learning workflows (supervised, unsupervised, self-supervised, shallow or deep learning), input image data are transformed by one or multiple distinct mathematical operations into a scientific result. The instructions for this transformation are learned from provided data (for example, labeled data for supervised learning and unlabeled data for unsupervised learning) to produce a machine learning model. However, the precise makeup of this model is not easily accessible to a user and depends strongly on the quality and nature of the supplied training data as well as on the specific training parameters. Biases in the training data and errors in the labels of ground truth for supervised machine learning will bias machine learning models73,74,75. Reporting is thus even more critical for reproducibility and understandability when machine learning applications are applied for image analysis.
Three major approaches are widely used in machine learning-based image analysis today that require different documentation. (1) Pretrained models are directly applied to new image data, and referral to existing references is sufficient. (2) Pretrained models are retrained (transfer learning) with new image data to improve the application, and, in this case, more information must be provided. (3) Models are trained de novo, in which case, extensive documentation is required for reproducibility.
Minimal (all models)
The precise machine learning method needs to be identifiable. Thus, the original method must be cited. At a minimum, access to the model that has been produced in the particular learning approach must be provided as well as validation input and output data. If a pretrained model has been used, it must be clearly identifiable. Example data or validation data must be provided; validation data must not be part of the training and testing data.
Recommended (retrained and new models)
To facilitate the reproduction and validation of results from either models trained from scratch or pretrained models that were retrained, the full training and testing data and any training metadata (for example, training time) should be made available. The code used for training the model should be provided. Code as well as data should be provided via public repositories (for example, Zenodo, GitHub). The authors should discuss and ideally test how well the model has performed and show any limitations of the used machine learning approach on their data. The application of machine learning models will particularly benefit from being deployed in a cloud-hosted format or via software containers.
Ideal (new models)
Further standardization promotes ease of reproduction and validation by the scientific community by making use of emerging online platforms. Thus, new models could be created conforming to standardized formats (for example, Model Zoo, https://bioimage.io/#/about) if they become more readily available in the future.
Discussion
Here, we have presented recommendations in the form of checklists to increase the understandability and reproducibility of published image figures and image analyses. While our checklists were initially intended for bioimages from light microscopes, we believe that their many principles are applicable more widely. Our checklists include recommendations for image formatting, annotation, color display and data availability, which, at the minimal level, can largely be achieved with commercial or open-source software (for example, ‘include scale bar’). Likewise, the minimal suggestions for image-analysis pipelines can be implemented readily with today’s options (for example, code repositories). We believe that, once included in microscopy core facility training and microscopy courses and introduced as guidelines from publishers, the recommendations will present no additional burden. On the contrary, transparent requirements for publishing images and progress-monitoring checklists will ease the path from starting a microscopy experiment to producing reproducible76, understandable image figures for all scientists.
Recommendations extending the ‘minimal’ level are introduced in the ‘recommended’ and ‘ideal’ reporting levels and at times go beyond what is easy to implement with standard tools today. They are meant to encourage continuous striving toward higher-quality standards in image publishing. Before all these advanced standards can become a new norm, technologies, software and research infrastructure must still be improved. At present, no image database is used widely enough to become a go-to solution, although dedicated resources exist and are slowly gaining traction and publishers are experimenting with parallel solutions (for example, EMBO source data). Also, while funding agencies increasingly require data to be deposited in repositories, few guidelines are provided for publishing terabytes to petabytes of raw data. While publishers may mandate data deposition or availability, they are not always reviewing its implementation. Combined with a lack of recognition of efforts put into publishing original image data, scientists are often discouraged to make data openly available. Commercial solutions for data storage are becoming increasingly available. For instance, Amazon Web Services Open Data has already been used to host image data (https://registry.opendata.aws/cellpainting-gallery/), and we believe that, ultimately, images presented in most publications should be linked to images processed by lossless compression that are amenable to reanalysis.
The checklists and recommendations for image analyses will naturally be dynamic and require regular updates to reflect new developments in this active research domain. Moreover, it is possible that generation of publication-quality images will also become a standardized ‘workflow’ in and of itself. It was previously suggested that images should be processed through scripting, with every step from microscope output to published figure stored in a metadata file39. Another challenge is the continuous availability of image-analysis software and workflows, which require software maintenance and updates to stay usable. Beyond technical developments, it is important to create inclusive standards for image publication that are achievable for our diverse global scientific community, which differs greatly in access to training and support, imaging infrastructure and imaging software. Our explicit intention is that the minimal level, which we believe must be met, does not pose an additional monetary or skill burden on scientists and is achievable with attention to detail. Our further reporting levels, ‘recommended’ and ‘ideal’, should encourage scientists to improve the accessibility and explanatory power of their images in publications.
We envision that the present checklists will be continuously updated by the scientific community and adapted to future requirements and unforeseen challenges. We are currently working on version 1.0 of a web-based Jupyter Book as a home for the ongoing development of ideas, extensions and also discussions on image publication. This consortium will continue future work in close alliance with similar initiatives such as NEUBIAS66,77, BINA and German BioImaging, initiatives that members of the authorship are involved with alongside their participation in QUAREP-LiMi. Collectively, we will work toward providing educational materials and tutorials based on the presented checklists and to continuously lobby to integrate its contents in general resources for better images78. We ask that all readers consider how their work will be seen and used in the future and join us in building a stronger scientific foundation for everyone. Version 1.0 of the presented checklists will already make images in publications more accessible, understandable and reproducible, providing a valuable resource as a solid foundation within today’s research that will benefit future science and scientists.
Data availability
The checklists can be downloaded as printable files from https://doi.org/10.5281/zenodo.7642559. The companion Jupyter Book can be found at https://quarep-limi.github.io/WG12_checklists_for_image_publishing/intro.html.
References
North, A. J. Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J. Cell Biol. 172, 9–18 (2006).
Brown, C. M. Fluorescence microscopy—avoiding the pitfalls. J. Cell Sci. 120, 1703–1705 (2007).
Senft, R. A. et al. A biologist’s guide to planning and performing quantitative bioimaging experiments. PLOS Biol. 21, e3002167 (2023).
Jonkman, J. Rigor and reproducibility in confocal fluorescence microscopy. Cytometry A J. Int. Soc. Anal. Cytol. 97, 113–115 (2020).
Heddleston, J. M., Aaron, J. S., Khuon, S. & Chew, T.-L. A guide to accurate reporting in digital image acquisition—can anyone replicate your microscopy data? J. Cell Sci. 134, jcs254144 (2021). This paper provides a nicely detailed breakdown of why complete reporting of methods in microscopy is important, who the stakeholders are and where the changes and motivation need to come from.
Montero Llopis, P. et al. Best practices and tools for reporting reproducible fluorescence microscopy methods. Nat. Methods 18, 1463–1476 (2021).
Hammer, M. et al. Towards community-driven metadata standards for light microscopy: tiered specifications extending the OME model. Nat. Methods 18, 1427–1440 (2021).
Rigano, A. et al. Micro-Meta App: an interactive tool for collecting microscopy metadata based on community specifications. Nat. Methods 18, 1489–1495 (2021). The Micro-Meta App demonstrates some of the steps being taken to provide real tools for scientists to use to improve microscopy methods reporting. It is not enough to simply scold scientists that something must change; rather, it is important that the tools to make such change as quick and painless as possible be created and made freely available.
Laissue, P. P., Alghamdi, R. A., Tomancak, P., Reynaud, E. G. & Shroff, H. Assessing phototoxicity in live fluorescence imaging. Nat. Methods 14, 657–661 (2017).
Kiepas, A., Voorand, E., Mubaid, F., Siegel, P. M. & Brown, C. M. Optimizing live-cell fluorescence imaging conditions to minimize phototoxicity. J. Cell Sci. 133, jcs242834 (2020).
Sheen, M. R. et al. Replication study: biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. eLife 8, e45120 (2019).
Jambor, H. et al. Creating clear and informative image-based figures for scientific publications. PLoS Biol. 19, e3001161 (2021). This study examined how effectively images conveyed insights in high-impact biology publications. It specifically focused on identifying the frequency of unclear images that lack crucial information such as scale bars, annotation legends or accessible colors and served as the catalyst for the current research project.
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22, 433 (2021).
Dietz, C. et al. Integration of the ImageJ ecosystem in the KNIME Analytics Platform. Front. Comput. Sci. 2, 8 (2020).
Perkel, J. M. Python power-up: new image tool visualizes complex data. Nature 600, 347–348 (2021).
Eliceiri, K. W. et al. Biological imaging software tools. Nat. Methods 9, 697–710 (2012).
Haase, R. et al. A hitchhiker’s guide through the bio-image analysis software universe. FEBS Lett. 596, 2472–2485 (2022).
Aaron, J. & Chew, T.-L. A guide to accurate reporting in digital image processing—can anyone reproduce your quantitative analysis? J. Cell Sci. 134, jcs254151 (2021).
Miura, K. & Tosi, S. in Standard and Super‐Resolution Bioimaging Data Analysis: a Primer (eds. Wheeler, A. & Henriques, R.) 269–284 (Wiley, 2017).
Ellenberg, J. et al. A call for public archives for biological image data. Nat. Methods 15, 849–854 (2018).
Hartley, M. et al. The BioImage Archive—building a home for life-sciences microscopy data. J. Mol. Biol. 434, 167505 (2022).
Williams, E. et al. The Image Data Resource: a bioimage data integration and publication platform. Nat. Methods 14, 775–781 (2017).
Ouyang, W. et al. BioImage Model Zoo: a community-driven resource for accessible deep learning in bioimage analysis. Preprint at bioRxiv https://doi.org/10.1101/2022.06.07.495102 (2022).
Boehm, U. et al. QUAREP-LiMi: a community endeavor to advance quality assessment and reproducibility in light microscopy. Nat. Methods 18, 1423–1426 (2021). This paper describes the network QUAREP-LiMi, in which this publication’s authors are embedded, and how their work is interconnected to the other QUAREP-LiMi working groups with related topics.
Nelson, G. et al. QUAREP-LiMi: a community-driven initiative to establish guidelines for quality assessment and reproducibility for instruments and images in light microscopy. J. Microsc. 284, 56–73 (2021).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Faklaris, O. et al. Quality assessment in light microscopy for routine use through simple tools and robust metrics. J. Cell Biol. 221, e202107093 (2022).
Bik, E. M., Casadevall, A. & Fang, F. C. The prevalence of inappropriate image duplication in biomedical research publications. mBio 7, e00809–16 (2016). This paper is a thorough quantitative and systematic analysis of image manipulations in publications. The paper has had a profound impact on scientific communities and highlighted the need to improve image quality.
Bik, E. M., Fang, F. C., Kullas, A. L., Davis, R. J. & Casadevall, A. Analysis and correction of inappropriate image duplication: the molecular and cellular biology experience. Mol. Cell. Biol. 38, e00309-18 (2018).
Cromey, D. W. Digital images are data: and should be treated as such. Methods Mol. Biol. 931, 1–27 (2013).
Editorial Policy Committee, Council of Science Editors. CSE’s Recommendations for Promoting Integrity in Scientific Journal Publications http://www.councilscienceeditors.org/wp-content/uploads/entire_whitepaper.pdf (2023).
Rossner, M. & Yamada, K. M. What’s in a picture? The temptation of image manipulation. J. Cell Biol. 166, 11–15 (2004).
Van Noorden, R. Publishers launch joint effort to tackle altered images in research papers. Nature https://doi.org/10.1038/d41586-020-01410-9 (2020).
Koppers, L., Wormer, H. & Ickstadt, K. Towards a systematic screening tool for quality assurance and semiautomatic fraud detection for images in the life sciences. Sci. Eng. Ethics 23, 1113–1128 (2017).
Bucci, E. M. Automatic detection of image manipulations in the biomedical literature. Cell Death Dis. 9, 400 (2018).
Van Noorden, R. Journals adopt AI to spot duplicated images in manuscripts. Nature 601, 14–15 (2022).
Martin, C. & Blatt, M. Manipulation and misconduct in the handling of image data. Plant Cell 25, 3147–3148 (2013).
Miura, K. & Norrelykke, S. F. Reproducible image handling and analysis. EMBO J. 40, e105889 (2021). This work demonstrates, with many examples, the importance of proper image analysis to avoid misleading images. The authors also make a strong case for creating reproducible figures with (for example, IJ-Macro) scripting.
Marqués, G., Pengo, T. & Sanders, M. A. Imaging methods are vastly underreported in biomedical research. eLife 9, e55133 (2020).
Nature Research Journals. Nature Guidelines https://www.nature.com/documents/nprot-guide-to-preparing-final-artwork.pdf (2023).
Schmied, C. & Jambor, H. K. Effective image visualization for publications—a workflow using open access tools and concepts. F1000Res. 9, 1373 (2020).
Cromey, D. W. Avoiding twisted pixels: ethical guidelines for the appropriate use and manipulation of scientific digital images. Sci. Eng. Ethics 16, 639–667 (2010). This article provides a first set of guidelines on how to properly treat digital images in scientific publications.
Russ, J. C. The Image Processing Handbook (CRC, 2006).
Zuiderveld, K. in Graphics Gems 474–485 (Elsevier, 1994).
Richardson, W. H. Bayesian-based iterative method of image restoration. J. Opt. Soc. Am. 62, 55–59 (1972).
Krull, A., Buchholz, T.-O. & Jug, F. Noise2Void—learning denoising from single noisy images. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2124–2132 (2019).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Fish, D. A., Brinicombe, A. M., Pike, E. R. & Walker, J. G. Blind deconvolution by means of the Richardson–Lucy algorithm. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 12, 58–65 (1995).
Crameri, F., Shephard, G. E. & Heron, P. J. The misuse of colour in science communication. Nat. Commun. 11, 5444 (2020).
Keene, D. R. A review of color blindness for microscopists: guidelines and tools for accommodating and coping with color vision deficiency. Microsc. Microanal. 21, 279–289 (2015).
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010).
Tedersoo, L. et al. Data sharing practices and data availability upon request differ across scientific disciplines. Sci. Data 8, 192 (2021). Data sharing and availability are crucial for reproducibility. This paper clearly documents how current data-sharing practices fall short and discusses ways to improve.
Gabelica, M., Bojčić, R. & Puljak, L. Many researchers were not compliant with their published data sharing statement: a mixed-methods study. J. Clin. Epidemiol. 150, 33–41 (2022).
Fisch, D. et al. Defining host-pathogen interactions employing an artificial intelligence workflow. eLife 8, e40560 (2019).
Erguvan, Ö., Louveaux, M., Hamant, O. & Verger, S. ImageJ SurfCut: a user-friendly pipeline for high-throughput extraction of cell contours from 3D image stacks. BMC Biol. 17, 38 (2019).
Klickstein, J. A., Mukkavalli, S. & Raman, M. AggreCount: an unbiased image analysis tool for identifying and quantifying cellular aggregates in a spatially defined manner. J. Biol. Chem. 295, 17672–17683 (2020).
Schmied, C., Soykan, T., Bolz, S., Haucke, V. & Lehmann, M. SynActJ: easy-to-use automated analysis of synaptic activity. Front. Comput. Sci. 3, 777837 (2021).
Tinevez, J.-Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Arganda-Carreras, I. et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017).
Arzt, M. et al. LABKIT: labeling and segmentation toolkit for big image data. Front. Comput. Sci. https://doi.org/10.3389/fcomp.2022.777728 (2022).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Berginski, M. E. & Gomez, S. M. The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res. 2, 68 (2013).
Hollandi, R. et al. nucleAIzer: a parameter-free deep learning framework for nucleus segmentation using image style transfer. Cell Syst. 10, 453–458 (2020).
da Veiga Leprevost, F. et al. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33, 2580–2582 (2017).
Cimini, B. A. et al. The NEUBIAS Gateway: a hub for bioimage analysis methods and materials. F1000Res. 9, 613 (2020).
Laine, R. F., Arganda-Carreras, I., Henriques, R. & Jacquemet, G. Avoiding a replication crisis in deep-learning-based bioimage analysis. Nat. Methods 18, 1136–1144 (2021).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer, 2015).
Falk, T. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018 (eds Frangi, A. F., Schnabel, J. A., Davatzikos, C., Alberola-López, C. & Fichtinger, G.) 265–273 (Springer, 2018).
Weigert, M., Schmidt, U., Haase, R., Sugawara, K. & Myers, G. Star-convex polyhedra for 3D object detection and segmentation in microscopy. 2020 IEEE Winter Conference on Applications of Computer Vision 3655–3662 (IEEE, 2020).
Moshkov, N. et al. Learning representations for image-based profiling of perturbations. Preprint at bioRxiv https://doi.org/10.1101/2022.08.12.503783 (2022).
Obermeyer, Z., Powers, B., Vogeli, C. & Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 366, 447–453 (2019).
Seyyed-Kalantari, L., Liu, G., McDermott, M., Chen, I. Y. & Ghassemi, M. CheXclusion: fairness gaps in deep chest X-ray classifiers. Pac. Symp. Biocomput. 26, 232–243 (2021).
Larrazabal, A. J., Nieto, N., Peterson, V., Milone, D. H. & Ferrante, E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl Acad. Sci. USA 117, 12592–12594 (2020).
Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016). This news article nicely summarizes the general problem that QUAREP-LiMi is attempting to handle within the field of microscopy. While the article covers a much broader scope, it provides an excellent overview of the impact of the reproducibility issue with clear graphics and a palatable length for any scientist.
Martins, G. et al. Highlights from the 2016–2020 NEUBIAS training schools for bioimage analysts: a success story and key asset for analysts and life scientists. F1000Res. 10, 334 (2021).
Collins, S., Gemayel, R. & Chenette, E. J. Avoiding common pitfalls of manuscript and figure preparation. FEBS J. 284, 1262–1266 (2017).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Jambor, H. et al. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife 4, e05003 (2015).
Sarov, M. et al. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068 (2016).
Cimini, B. A comparison of repositories for deposition of light microscopy data. Zenodo https://doi.org/10.5281/zenodo.7628604 (2023).
Acknowledgements
We acknowledge the support of and discussions with all our colleagues and QUAREP-LiMi Working Group 12 members. We additionally thank O. Biehlmeier, M. Bornhäuser, O. Burri, C. Jacobs, A. Laude, K. Ho and R. D’Antuono for further feedback and discussions on the manuscript and their endorsement of the checklists. Icons were designed by the authors and are from https://fontawesome.com; ImageJ sample images are from refs. 79,80,81. C.S. was supported by a grant from the Chan Zuckerberg Initiative napari Plugin Foundation (2021-24038). C.B. was supported by grants from ANID (PIA ACT192015, Fondecyt 1210872, Fondequip EMQ210101, Fondequip EQM210020) and PUENTE-2022-13 from Pontificia Universidad Católica de Chile. B.A.C. was funded by NIH P41 GM135019 and grant 2020-225720 from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation. F.J. was supported by AI4Life (European Unions’s Horizon Europe research and innovation program; 101057970) and by the Chan Zuckerberg Initiative napari Plugin Foundation (2021-240383 and 2021-239867). M. Hammer and D.G. were supported by NSF award 1917206 and NIH award U01 CA200059. R.N. was supported by grant NI 451/10-1 from the German Research Foundation and grant 03TN0047B ‘FluMiKal’ from the German Federal Ministry for Economic Affairs and Climate Action. A.P.-D. was supported by EPSRC grant EP/W024063/1. C.S.-D.-C. was supported by grant 2019-198155 (5022) awarded by the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, as part of their Imaging Scientist Program. She was also funded by NIH grant U01 CA200059. C.T. was supported by a grant from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation (grant 2020-225265). H.K.J. was supported by MSNZ funding of the Deutsche Krebshilfe.
Author information
Authors and Affiliations
Contributions
C.S., conceptualization, methodology, writing original draft, visualization, supervision, project administration; M.S.N., conceptualization, methodology, writing original draft, visualization, writing (review and editing); S.A., endorsement and reviewing; G.-J.B., endorsement and reviewing; C.B., critical review (commentary in pre-writing stage), review and editing; J. Bischof, endorsement and reviewing; U.B., conceptualization, methodology, writing (review and editing), endorsement; J. Brocher, endorsement, writing (review and editing), visualization; M.T.C., conceptualization, methodology, endorsement, writing (review and editing); C.C., endorsement and reviewing; J.C., writing (review and editing); B.A.C., writing (review and editing), endorsement; E.C.-S., endorsement and reviewing; M.E., writing (review and editing); R.E., endorsement and reviewing; K.E., endorsement and reviewing; J.F.-R., endorsement and reviewing; N.G., endorsement and reviewing; L.G., endorsement and reviewing; D.G., writing (review and editing); T.G., writing (review and editing); N.H., endorsement, review and editing; M. Hammer, review and editing; M. Hartley, endorsement and reviewing; M. Held, endorsement and reviewing; F.J., endorsement and reviewing; V.K., writing (review and editing); A.A.K., endorsement, reviewing and editing; J.L., endorsement and reviewing; S.L.D., endorsement and reviewing; S.L.G., writing (review and editing); P.L., endorsement and reviewing; G.G.M., conceptualization, methodology, writing (review and editing); A.M., reviewing and editing, endorsement; K.M., conceptualization, methodology, writing (review and editing); P.M.L., endorsement and reviewing; R.N., conceptualization, supervision, project administration, endorsement; A.N., endorsement and reviewing; A.C.P., conceptualization, methodology, writing (review and editing); A.P.-D., writing (review and editing), visualization; L.P., writing (review and editing); R.A., endorsement and reviewing; B.S.-D., endorsement and reviewing; L.S., endorsement and reviewing; R.T.S., endorsement and reviewing; A.S., review and editing; O.S., endorsement, writing (review and editing); V.P.S., endorsement, reviewing; M.S., endorsement and reviewing, writing (review and editing); S.S., resources, writing (review and editing); C.S.-D.-C., conceptualization, endorsement, validation; D.T., writing (review and editing); C.T., conceptualization, methodology, writing (review and editing); H.K.J., conceptualization, methodology, writing original draft, visualization, project administration, supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Gary Laevsky, Jennifer Peters and Brian Slaughter for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Scientific repositories.
Overview of current repositories that accept image data82.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schmied, C., Nelson, M.S., Avilov, S. et al. Community-developed checklists for publishing images and image analyses. Nat Methods 21, 170–181 (2024). https://doi.org/10.1038/s41592-023-01987-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-023-01987-9